Translate this page into:
Protocol for a Longitudinal Analysis of the Vaginal Microbiome from a Pregnant Cohort of African Women in Nigeria
*Corresponding author email: nkechiodogwu27@gmail.com
-
Received: ,
Accepted: ,
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 Unported, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Background:
The vaginal microbiota is an important component of the reproductive health of women as it offers protection against urogenital infection. African women are reported to have a vaginal microbiota colonized with high proportions of strict anaerobes rather than lactobacillus- dominated microbes. These strict anaerobes have been associated with pre-term birth and neonatal disease. The prevalence of pre-term birth (PTB) in Africa poses a major challenge to reproductive healthcare, hence the clinical and scientific attention focused on understanding the causative mechanisms of PTB. A pragmatic approach to curbing PTB requires the identification of the vaginal microbiome during various stages of a healthy pregnancy (the ‘normal’). This information will provide baseline data for future investigations of vaginal microbiome that may cause PTB (the ‘abnormal’). We present a protocol for the longitudinal analysis of vaginal microbiome in a cohort of pregnant women in Southwest Nigeria.
Methods:
We propose to recruit 51 pregnant Nigerian women, enrolling them into the study at 17-21 gestational weeks. Two vaginal swab samples and three milliliters of blood would be collected at enrollment. Sample collection will be repeated at 27-31 weeks’ gestation, ≥36 weeks’ gestation, 24-48 hours after birth and 6 weeks post-partum. DNA will be extracted from the vaginal samples and 16S rRNA sequencing would be performed. Blood samples collected would be assayed by ELISA technique for placental steroid hormones. Data will be statistically analyzed and considered in the light of vaginal microbial diversity, clinical, nutrition and other health data.
Conclusion and Global Health Implication:
Our data set will bring new insights into the vaginal microbiome of apparently healthy African women in pregnancy and postpartum, which should serve as a baseline for the investigation of vaginal microbes that may provide useful information for the prediction and management of preterm birth. It is anticipated that these data will facilitate future personalized therapeutic management and consequently improve the reproductive health fitness of women in Africa.
Keywords
Vaginal microbiome
Pregnancy
Pre-term Birth
Nigeria
Women
Longitudinal analysis
Protocol
1. Background
Public health initiatives to uncover the roots of, and address the persistent health disparities between people of African descent and others continue to face challenges. Notable is the rate of pre-term birth (PTB) and neonatal outcomes among African women and people of African descent in the diaspora, compared to their Caucasian counterparts. Compared to their white counterparts, black women experience greater risk of birth before 37 completed gestational weeks (13.2% vs. 8.9%), lower birth weight (<2.5 kg) (13.2% vs. 7.0%), and higher infant mortality (10.81% vs. 5.07%).1 Faced with these continuing challenges, clinical and scientific attention has focused on understanding the underlying risk factors or the causal mechanisms for PTB.
Risk factors in black women that have been linked to adverse birth outcomes include race and bio-ethnicity,2 dietary threats,3 sexual practices,4 hygiene behavior,5 and smoking.6 The Vaginal microbiome signature during pregnancy has been shown to influence susceptibility to adverse pregnancy outcomes.7,8,9,10 Ravel et al.2 broke down the vaginal microbiome into 5 community state types (CST); CST I, Lactobacillus crispatus; CST II, Lactobaccillus gasseri; CST III, Lactobaccillus iners; CST V, Lactobaccilus jensenii; and CST IV, a mixture of Bacterial Vaginosis Associated Bacteria (BVAB) and other strict anaerobes.2
Several findings have attributed this disparity in PTB burden and adverse neonatal outcomes among Black and White women to the dominance of Bacterial Vaginosis Associated Bacteria (BVAB) or CST IV microbes in the vaginal niche of black women.11,12,13 With the advent of next-generation sequencing technology, deciphering the composition of the vaginal microbiome is now possible.14,15 An approach to understanding the vaginal microbiome signature in PTB (i.e., the ‘abnormal’) in our population is to first unravel the composition of the vaginal microbiome in ‘normal’ pregnant women. This approach will provide an opportunity to determine vaginal microbiomes that modulate and enhance pregnancy, differentiated from those that may predispose to PTB. Differentiating the vaginal microbial composition in healthy pregnancy from vaginal microbes involved in PTB will guide clinicians appropriately and allow personalized targeted intervention to prevent PTB and its associated adverse pregnancy outcomes.
In Africa, the vaginal microbiome composition has been studied in non-pregnant cohorts of women as reported by Anahtar et al.16 and Gosmann et al.17 in South Africa, by Borgdorff et al.18 and Jesper et al.19 in Rwanda, by Lokken et al.20 in Kenya, by Frank et al. in Burkina Faso21 and by Anukam et al. in Nigeria.22 However, to our knowledge, no prior study has described the vaginal microbiome composition in pregnant Africans or those with PTB. Pregnancy is accompanied by an increased dominance in Lactobacillus species which enhances stability.23,24 However, this situation changes postpartum,7 an alteration suggested to be driven by estrogen.25,26 In this study protocol, we propose to follow-up recruited participants after delivery and have their samples collected at 6 weeks postpartum (the puerperium) so as to describe the association of pregnancy-associated hormones with the vaginal microbiota at the puerperium. This is an aspect where other studies done in non-African populations have been limited. Our study will focus on exploiting the 16S rRNA sequencing to characterize the vaginal microbiome. The basis of this approach is the opportunity it affords to determine unculturable microbes in the vaginal niche during pregnancy. It will also improve our understanding of the vaginal microbiome in healthy pregnancy, providing baseline data for future investigations of microbes associated with PTB.
The objectives of this study are to: (1) characterize the vaginal microbiome at 17-21 weeks, 27-31 weeks, ≥ 36 weeks’ gestation, and at 24-48 hours and 6 weeks postpartum by 16S rRNA sequencing; (2) describe any association between plasma estrogen and progesterone with changes in the vaginal microbiome; and (3) evaluate the association of the vaginal microbiome with other clinical parameters such as previous pregnancy history, pregnancy outcomes, birth outcomes, and other biobehavioral factors. This paper presents the research design, data collection plan, and the proposed laboratory methods.
2. Methodology
2.1. Study Area and Design
This prospective longitudinal descriptive study’ would be conducted at the University college Hospital (UCH), Ibadan, Nigeria. UCH is a tertiary-level hospital located in South-western Nigeria that serves patients from diverse social and economic status and from different states in Nigeria. Located in Ibadan, the largest city by land area in West Africa, UCH has community-based outreach services where primary and secondary healthcare services are provided. The hospital has more than 60 service and clinical departments and runs 96 consultative outpatient clinics per week in 50 specialty and sub-specialty disciplines. There are about 1,000 inpatient beds. The UCH Obstetrics and Gynecology Department handles antenatal and postnatal patient care, emergencies and referrals. The clinic operates daily (4 days for antenatal clinics and 1 day for the postnatal clinic). On every antenatal clinic or post-natal clinic day, an average of 90 patients are seen.
2.2. Conceptual Framework
This study is guided by a conceptual framework (Figure 1). This framework postulates that nutrition/diet, sex, behavioral health practice, circulating levels of estradiol and progesterone all impact the vaginal microbiome directly. Based on this conceptual framework, we would expect that the vaginal microbiome consequently may promote or exclude pathogens, thus influencing maternal and neonatal outcomes.
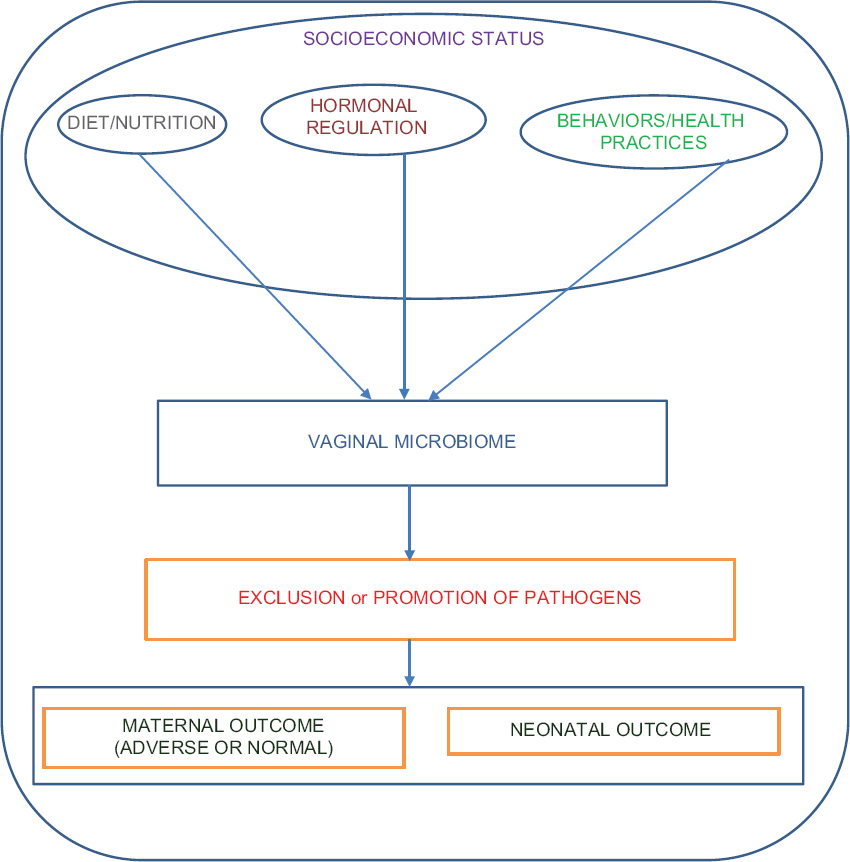
- Conceptual framework
2.3. Study Population and Sites
This study will be conducted at UCH, Ibadan, Nigeria. Nigeria lies on the west coast of Africa between latitudes 4º16’ and 13º53’ north and longitudes 2º40’ and 14º41’ east and is the most populous country in Africa and the 14th largest in land mass.27 Nigeria features over 500 ethnic groups, six geopolitical regions, different languages and an estimated population of over 200 million, ranking 7th in the world.28 UCH is one of the 24 teaching and research federal teaching hospitals in Nigeria where people can access health care services within their reach.
2.4. Participant Recruitment
Healthy pregnant women will be recruited at first time-point between 17 and 21-weeks’ gestation. Table 1 shows the study’s inclusion and exclusion criteria.
| Inclusion criteria | Exclusion criteria |
|---|---|
| Participants will be included if they are: 1) Between 17- and 21 weeks’ gestation (confirmed by clinical records and ultrasound result) and with singleton pregnancy; 2) Women within reproductive age (18-49 years); 3) No known pregnancy complications at the first obstetric visit; 4) Able to provide written informed consent, willing to participate in all aspects of the study; and 5) Weigh greater than 110 pounds (50 kilograms), a standard requirement in obstetric studies that include blood draws. |
Participants will be excluded if they: 1) Had sexual activity within 72 hours of sampling; 2) Reported vaginal bleeding in the preceding week; 3) Used antibiotics in the preceding 2 weeks; 4) Had chronic active viral infections, including HIV-1/2, HTLV-1/2, hepatitis B/C; and 5) Have a known autoimmune disease, such as rheumatoid arthritis or systemic lupus erythematosus; solid organ or transplant recipient. |
2.5. Data Collection
Table 2 presents the data that will be collected at five time-points of the study. These include: three times during pregnancy (17-21 weeks, 27-31 weeks, 36 weeks and above) and two times at 24-48 hours and at 6 weeks postpartum. Sociodemographic data such as age, level of education, marital status, employment, family size and insurance status will also be collected.
| Clinical data | Health survey | Diet and nutrition |
|---|---|---|
| Pregnancy BMI: Measured patient height and weight according to the standard acceptable description.
30 Pregnancy outcome such as early miscarriage, late miscarriage, preterm delivery, term delivery, gestational age at birth and delivery type. |
Diagnosis of genital disease before and in pregnancy. Use of antibiotics (oral or systemic). Use of topical medication on the vagina Sexual encounters (type and frequency of intercourse. Use of condoms. Number of sexual partner per lifetime). Hygiene self-care practices (use of disinfectant and antiseptics on vaginal surface, douching and feminine sprays/wipes on vagina). Smoking of tobacco. Parity status. Outcome of previous pregnancy. Complaints during pregnancy. type of vaginal discharge will be obtained |
Food Frequency Questionnaire (FFQ) will be used to register all diet and supplement taken by participants during pregnancy.31 The FFQ will be modified to include: Intake of Essential trace elements, vitamins D and B, folates; Intake of probiotic feed; Intake of probiotic supplement; and Type of diet (local/traditional diet or modern diet). |
2.6. Bioassays and Sample Processing
I. Vaginal Gram stain: At the microbiology laboratory, vaginal swabs will be smeared unto slides. Smeared slides will be dried, heat-fixed and gram staining procedures done using the manufacturer’s protocol. The slides will be scored by using Nugent criteria.29 High scores will be classified as 7-10, Intermediate as 4-6, and low scores as 0-3.
II. Hormonal Assay: Blood samples will be centrifuged at 3000 rpm for 15 minutes. Thereafter, plasma will be decanted and estradiol and progesterone will be assayed using ELISA according to the manufacturer’s protocol.
III. DNA Extraction and 16S r RNA Gene sequencing
DNA extraction will be done in line with standardized protocol for the Human Microbiome project.32 The integrity of the extracted bacterial DNA will be confirmed by PCR amplification using universal primers. Polymerase chain reaction will be done with template DNA. Sequencing of the 16S rRNA gene will be done with established protocol.14
2.7. Data Management and Storage
Questionnaire and clinical data will be directly entered into computer by the principal investigator. As storage/back up plan all data will also be documented in a notebook and stored in the personal computer of the principal investigator and will be backup twice in personal external backup devices. We anticipate that study will be completed within 24 months. Table 3 outlines the proposed activities for each time period.
| Activities | Time frame |
|---|---|
| Recruiting 51 participants from (17 weeks to 6 weeks postpartum) | 11 months |
| DNA extraction | 2 months |
| Estimation of hormones concentration via ELISA technique | 1 month |
| DNA Sequencing | 2 months |
| Bioinformatic analysis of sequence data | 2 months |
| Statistical analysis | 2 months |
| Data Interpretation and compilation of results | 4 months |
2.8. Statistical Plan
2.8.1. Sample size determination
Our proposed sample size of 51 women after a 5% attrition will provide sufficient dataset to enable the comparison of microbiome characteristics between groups of women at varying gestational ages during pregnancy and in the puerperium.
We calculated our sample size using the formula described below
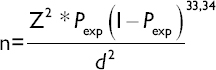
Where: Z= level of confidence according to the standard normal distribution (for a level of confidence of 95%, Z = 1.96; n = required sample size; Pexp = expected prevalence; and d = desired absolute precision at 5%=0.05.
For metagenomic studies, the Shannon diversity index is used to characterize species diversity in a community, depending on the community state type of microbes found in the vaginal niche.35 Considering a 20% prevalence of CST IV associated with the VMB of women of African descent.10 We substituted Pexp and other variables in the formula below:
N= (1.962) (0.2) (0.8)/(0.05)2 = 246; Adjusting for attrition (5%), N = (246)/(0.95) =259. Recruited participants will be sampled at 5 time points (259/5) =51.8. Therefore, samples will be collected from 51 pregnant women at 5 timepoints.
2.8.2. Data Analysis Methods
2.8.3. Statistical analyses
a) To determine statistical differences between the vaginal microbiome throughout gestation and the puerperium, the Statistical Analysis of Metagenomics Profile (STAMP) software package would be used. Data will be subjected to multivariate analysis in the form of principal component analysis (PCA) to obtain variance that correlated in the dataset.
b) To obtain the statistical significance of microorganism abundance and community state types (CST) during pregnancy and at the puerperium, a linear mixed model regression analysis would be used.
c) To measure if there is a significant change in bacterial species abundance during pregnancy, linear regression analysis will be done for each microorganism.
d) To investigate if there is a significant change in microorganism abundance after pregnancy, a binary time variable will be created for measurements during pregnancy and also for postpartum measurements. Using this binary time variable a linear regression for each microorganism will be performed.
e) To determine if CSTs change significantly over the course of pregnancy a linear regression for each CST will be performed.
f) To test if CSTs change significantly during the puerperium, a binary time variable would be created with time for measurements during pregnancy and time for postpartum measurements and linear regression performed for each CST.
g) To describe the association between the composition of the vaginal microbiome at pregnancy and the postpartum and hormonal regulation, a bivariate analysis will be done. The logistic regression model will also be used to test the significance of the interaction between other biobehavioral factors, sociodemographic factors and the microbiome compositions.
3. Discussion
In this paper, we present the design data collection methods and expected statistical plan to achieve our study objectives. The strength of our study includes the opportunity to decipher the vaginal microbiome signature during pregnancy and describe the association of the vaginal microbiome during pregnancy with pregnancy-associated hormones. The study also provides the opportunity to collate high quality clinical data (three time points during pregnancy, 24-48 hours post-delivery, and at 6 weeks post-partum) and other health variables. Thirdly our detailed questionnaire will highlight both socio-demographic, health and behavioral characteristics, which are also important factors that determine the vaginal community.
Interestingly, the study seeks to exploit 16S rRNA gene sequencing, an advanced technique, against an organism-centric approach generally used for bacterial identification in sub-Saharan Africa. We envisage that an area of weakness in the study is the veracity of what the women report in the questionnaire. Women are usually reserved in discussing their past and current sexual experiences and may provide incorrect information.36,37 Unless, there are health complications, most women in sub-Saharan Africa are unlikely to disclose their early sexual debut and tobacco intake for fear of social stigma. Details like a woman’s smoking (details on number of cigarettes per day) and sexual behavior (anal or oral sex; number of lifetime sexual partners; frequency of sexual intercourse) may not get accurate responses.
The impact of our study extends beyond just characterizing the vaginal microbiome during pregnancy. The data collected will probably trigger the asking of other research questions as well as either proving or refuting some existing hypotheses. We present this study as a baseline study and envisage that, through similar studies in pregnant women in other centers in Nigeria and Africa, we would be able to more easily detect imbalances of the vaginal microbiome. This would allow the development of more personalized and targeted interventions in order to restore a microbiome that optimizes reproductive health in individual women, especially pregnant and post-natal women in Nigeria. Ultimately, we hope this study will lead to significant breakthroughs in the reproductive health and content of care in women in Nigeria, Africa and around the world.
4. Conclusion and Global Health Implications
Globally, it has been the goal of African governments to reduce preterm births. While several developed countries had made giant strides and effort to reduce its prevalence, the rate remains stubbornly higher among people of African descent. Our data set will bring new insights into the vaginal microbiome of apparently healthy African women during pregnancy and postpartum in a Nigerian population. This should serve as a baseline for the investigation of vaginal microbes that may contribute to preterm birth. It is anticipated that these data will facilitate future personalized therapeutic management and consequently improve the reproductive health fitness of women in Africa. This impact will also have economic advantage as expenses for parental care and medical management for premature babies will also potentially reduce.
Acknowledgement:
The unalloyed support of Pan African University (PAU), of the African Union Commission is highly acknowledged especially for providing NMO a platform as a PhD student in Reproductive Biology Sciences. The authors would like to thank the anonymous reviewers for helpful suggestions, which resulted in a better overall paper.
Compliance with Ethical Standards
Competing Interest: The authors have no competing interests to disclose regarding the publication of this paper.
Funding: None.
Ethics approval: Study protocol was approved by the Ethics Board at the University College Hospital, Ibadan, Nigeria.
Author contributions: NMO conceptualized and designed the study, drafted and wrote the paper. NMO, CACO, OOO and AOO developed the study design, wrote and revised the article before final submission.
References
- Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4680-4687.
- [Google Scholar]
- Reprod Sci. 2012;19(9):939-948.
- Rapid fluctuation of the vaginal microbiota measured by Gram stain analysis. Sex Transm Infect. 2010;86:297-302.
- [Google Scholar]
- Determinants of excessive gestational weight gain in urban, low-income women. Women's Health Issues. 2012;22(5):e439-46.
- [Google Scholar]
- Prevalence and risk factors of bacterial vaginosis during the first trimester of pregnancy in a large French population- based study. Eur J Obstet Gynecol Reprod Biol. 2012;163(1):30-34.
- [Google Scholar]
- The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:8988.
- [Google Scholar]
- Recent advances in understanding the microbiology of the female reproductive tract and the causes of premature birth. Infect Dis Obstet Gynecol 2010:737425.
- [Google Scholar]
- The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18.
- [Google Scholar]
- Diversity of the vaginal Microbiome correlates with preterm birth. Reprod Sci. 2014;21(1):32-40.
- [Google Scholar]
- The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands. PLoS ONE. 2017;12(7):e0181135.
- [Google Scholar]
- The Vaginal Microbiome Consortium. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology. 2014;160(10):2272-82.
- [Google Scholar]
- Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. International Society of Microbial Ecology Journal. 2007;1(2):121-33.
- [Google Scholar]
- Nature. 2012;486:215-21.
- Kacey Eichelberger. Maternal microbiome and pregnancy outcomes. Fertility and Sterility. 2015;104(6):1358-1363.
- [Google Scholar]
- Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity. 2015;42:965-76.
- [Google Scholar]
- Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. 2017;46:29-37.
- [Google Scholar]
- Lactobacillus-dominated cervicovaginal microbiota associated with reduced HIV/STI prevalence and genital HIV viral load in African women. ISME J. 2014;8:1781-93.
- [Google Scholar]
- and the Vaginal Biomarkers Study Group. The significance of Lactobacillus crispatus and L vaginalis for vaginal health and the negative effect of recent sex:a cross-sectional descriptive study across groups of African women. BMC Infect Dis. 2015;15:115.
- [Google Scholar]
- Association of recent bacterial vaginosis with acquisition of mycoplasma genitalium. Am J Epidemiol. 2017;186:194-201.
- [Google Scholar]
- Altered vaginal microbiota are associated with perinatal mother-to-child transmission of HIV in African women from Burkina Faso. J Acquir Immune Defic Syndr. 2012;60:299-306.
- [Google Scholar]
- Vaginal bacteriome of Nigerian women in health and disease:A study with 16S rRNA metagenomics. Trop J Obstet Gynaecol. 2019;36:96-104.
- [Google Scholar]
- A metagenomic approach to characterization of the vaginal Microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466.
- [Google Scholar]
- Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L.gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiology. 2009;9:116.
- [Google Scholar]
- bold>Influence of vaginal bacteria and D- and L-lactic acid isomers on vaginal extracellular matrix metalloproteinase inducer:implications for protection against upper genital tract infections. mBIO. 2013;4(4):e00460-13.
- [Google Scholar]
- Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect Immun. 1998;66:1985-9.
- [Google Scholar]
- Nigeria Demographic and Health Survey. Abuja, Nigeria, and Rockville, Maryland, USA: NPC and ICF; 2018.
- Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol. 1991;29(2):297-301.
- [Google Scholar]
- Body Mass Index:Obesity, BMI, and Health:A Critical Review. Nutrition Today. 2015;50(3):117-128.
- [Google Scholar]
- Use of a food frequency questionnaire in American Indian and Caucasian pregnant women:a validation study. BMC Public Health. 2005;5:135.
- [Google Scholar]
- Manual of Procedures for the Human Microbiome Project. Washington, DC: National Institutes of Health; 2010. https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/GetPdf.cgi?id=phd002854.2
- Veterinary Epidemiology (3rd ed). Ames, IA: Blackwell Science; 2005.
- 2016. Research Aids on sample size formula for sample size calculation:Survey System. https://www.surveysystem.com/sample-size-formula.html
- The Mathematical Theory of Communication. Urbana, IL: University of Illinois Press; 1949.
- Questionnaires to measure sexual quality of life. Qual Life Res. 2004;13:1643-1658.
- [Google Scholar]
- Sexual life:A Clinician's Guide. New York, NY: Plenum Press; 1992.





